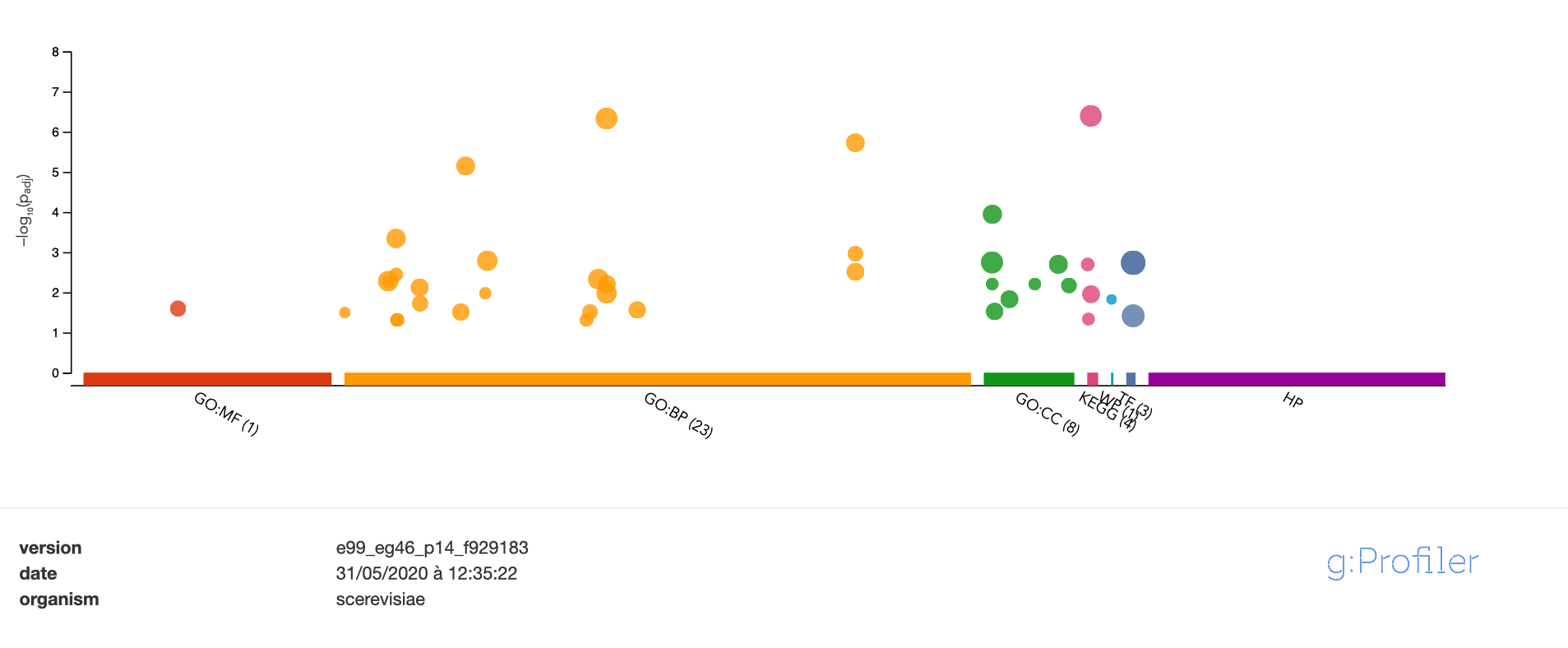
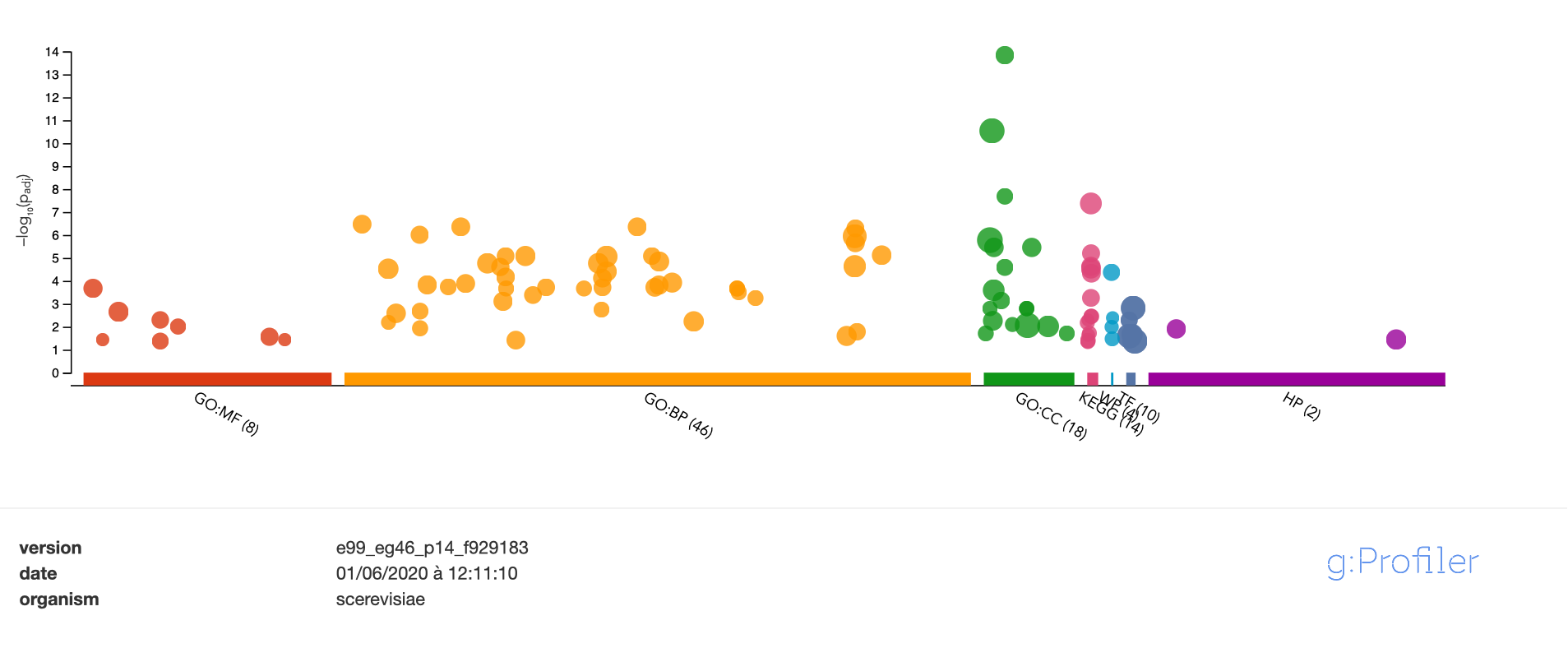
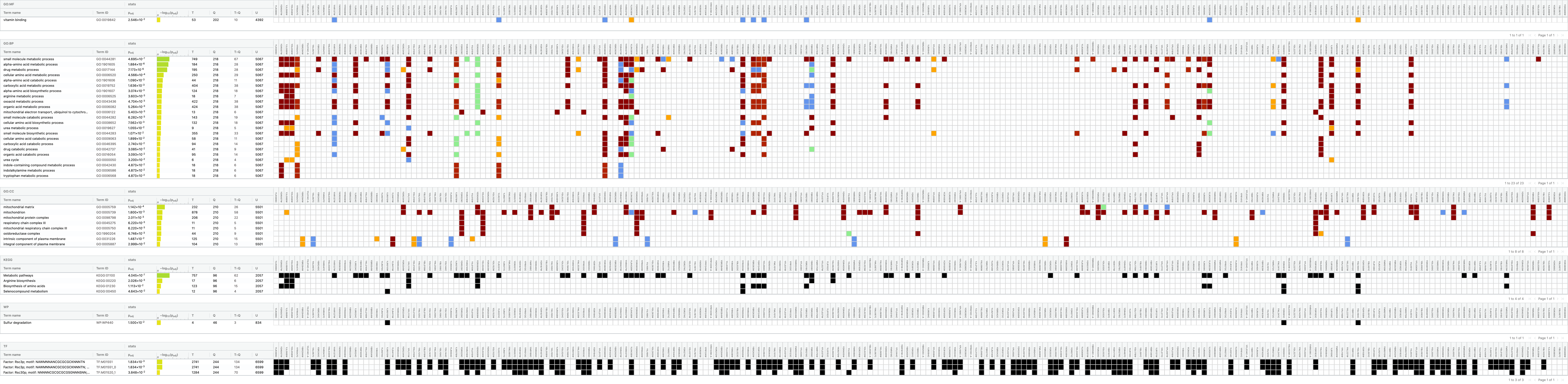
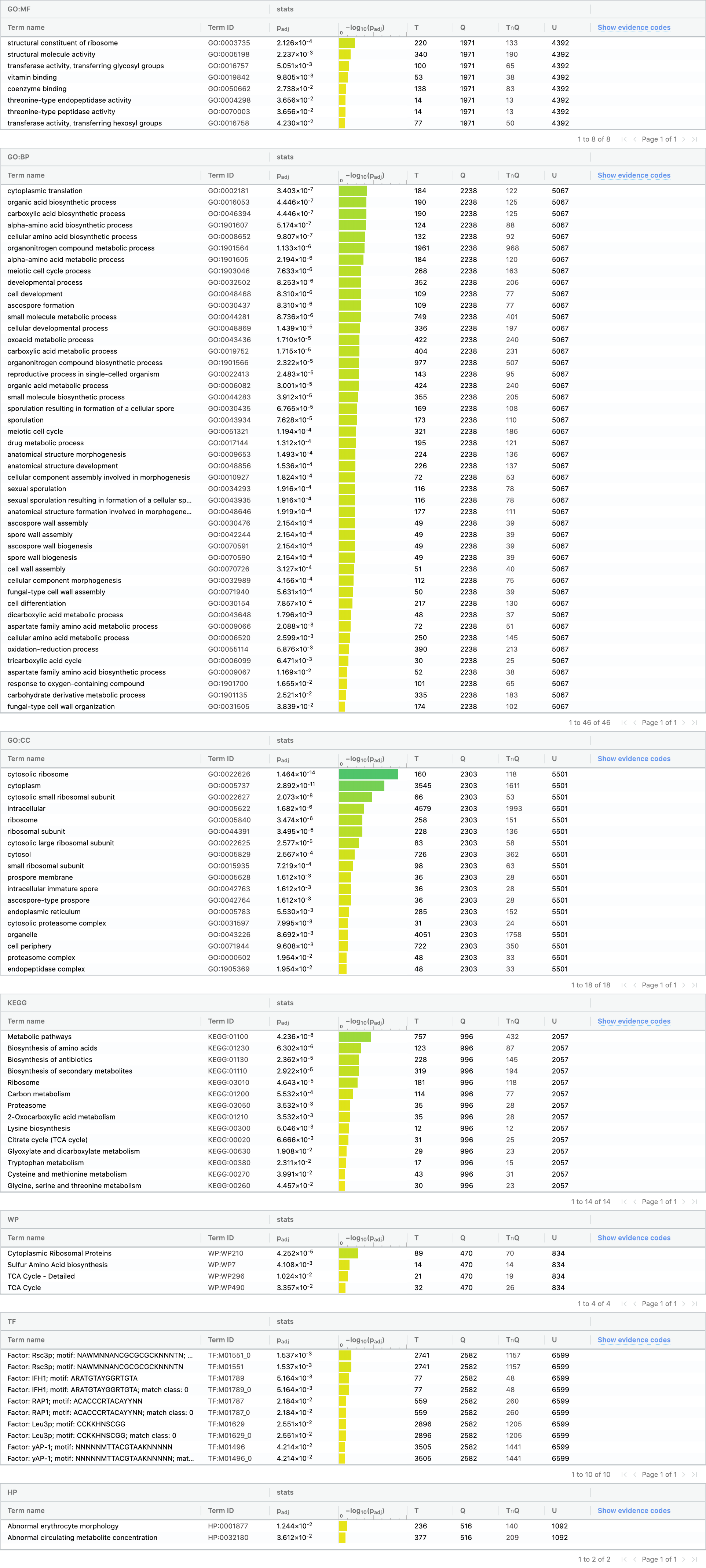
Gene set comparison (over-representation of the intersection)
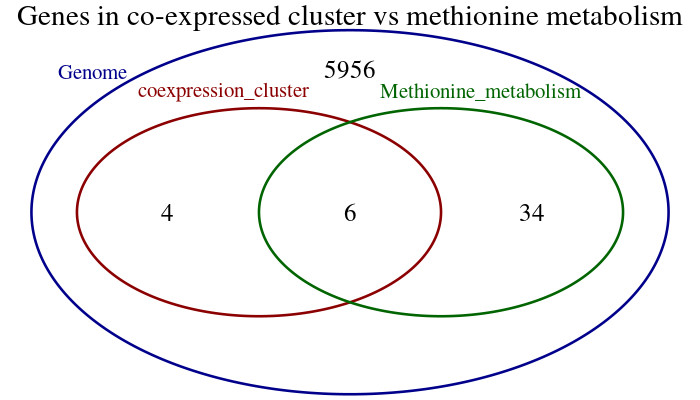
- A given organism has 6000 genes, 40 of which are involved in methionine metabolism.
- A set of 10 genes were reported as co-expressed in an RNAseq experiment. Among them, 6 are related to methionine metabolism.
- How significant is this observation ? More precisely, what would be the probability to observe such a correspondence by chance alone ?